SEQUAL is a spesific way of arranging a couple of algorithms and scripts in such a way that a quality label is generated for a given DNA sequencing trace file. For more informaion please visit http://timewarping.org/sequal or consult the README files.
We used a subset of DNA trace files freely available for noncommercial use from InSNP. We converted them to Standard Chromatogram Format (SCF) by using Phred. We didn't include the trace files because of legal issues. However The manually created quality labels of the SCF files can be found in data/labels.txt.
The SCF files are transformed into a set of features by using the MATLAB script src/createfeatures.m The features as well as the quality labels of the SCF file are saved into src/features.arff.
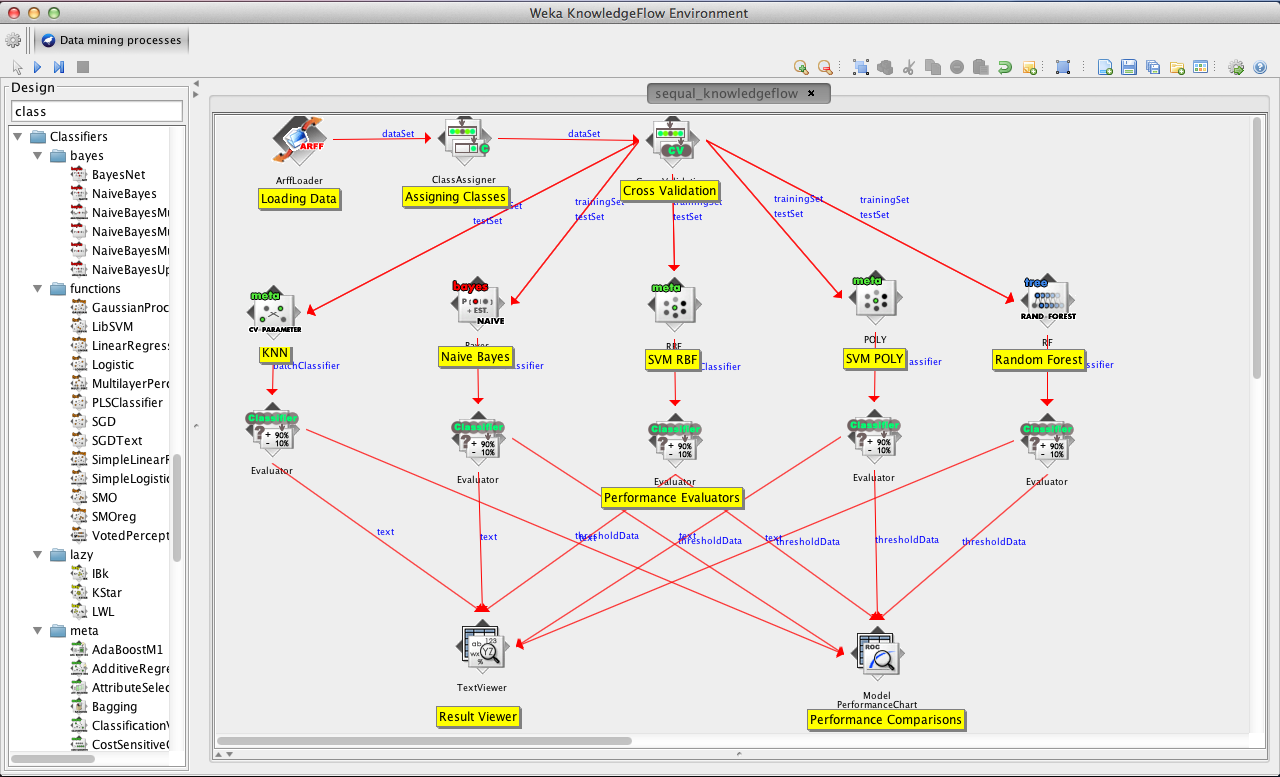
In order to classify the data with different machine learning algorithms, KnowledgeFlow Environment of WEKA is used. The KnowledgeFlow script is saved to src/knowledgeflow.kfml